Midbody and exosome research are hot topics right now! There is even an entire journal devoted to them and other subcellular particles: Journal of Extracellular Vesicles. Lots and lots of papers on this! Check out my models that demonstrate a connection between midbodies, exosomes, and cellular differentiation!
How TADs can communicate with each other and alter gene expression via enhancer connections based upon my models for DNA replicons.
a
“BACTERIA” MADE US WHO WE ARE TODAY?
Did you know Archaea have circular chromosomes?
s
s
https://www.bitchute.com/video/swj74z3yZk0e
If you work with tissue culture cells, try out my protocol and see what you get!
In 1988, I graduated with my PhD at Ohio State University. The title of my thesis is “Eukaryotic DNA Superstructure”. I continued my research at OSU and discovered something so amazing that it blew our collective minds. I had stained cells from mice to illuminate their DNA content. Most researchers would have expected to see brightly stained strands of chromatin, the stuff that makes up chromosomes. I saw the same exact thing too, until I chemically teased the chromatin apart. That’s when I discovered brightly illuminated, beaded circles in a wide variety of sizes. This was a novel and controversial finding that I thought would spark further research. But the scientific review process was as painful as it was illuminating. Nobody contacted me for further inquiry, not even today!
So I am reaching out to my fellow scientists, hoping to stimulate discussion, share information and, ideally, encourage additional scientific research. Is this discovery a biological sea change in understanding? Can the discovery process be reproduced? Could cancer cures be just around the corner, if we understood chromosome structure better than we do now? Do these questions warrant further investigation? Without additional research, we will never know. Contact me at fabernathy@sbcglobal.net.
The classic linear DNA model for human chromosomes violates the second law of thermodynamics
HOW CAN ANIMAL CHROMOSOMES BE PUT TOGETHER VIA ENDOSYMBIOSIS?
How Acid Hydrolysis Revealed the Circular Nature of Eukaryotic DNA
Video, Powerpoint
When it Comes to Chromosomes, You Have Been Misled
(Bitchute Channel)
Click here to check out the latest posts on this blog
This is actually two blogs in one
Blog #1 can be found under the page tab: General Posts. This is the one you may want to check out first. I update the blog here whenever I discover new exciting information.
Blog #2 is about the fallacies associated with the study of science and how they interfere with the general pursuit of knowledge and discovery. These are under the page tab Rantings of a Mad Scientist.
The biological models tab contains a list of reference links relating to the models that is periodically updated.
Throughout Blog #1, you will find a variety of microphotographs, some of them repeated for emphasis. Here is an example of one of them:

(click image to enlarge)
It’s pretty, mysterious, and full of little beaded circles that appear to be emanating from a larger “mother ship”. At least, that’s how my Ph.D. advisor described them at Ohio State University almost 25 years ago. These were actually discovered after I had already completed my Ph.D., so you won’t find them in my dissertation or any subsequent manuscripts. They came out of mouse cells that were in the process of dying under the microscope. The green color is due to fluorescence from a dye the cells were exposed to that allows nucleic acids to light up under this kind of microscope. I managed to “coax” them out of dying cells using dilute acid that partially degrades the DNA.
If you are a DNA biologist, you may want to skip down to the part where I “boil” the blog down regarding origins of replication, promoters, enhancers, and splice sites.
Since I have no idea of your level of mastery of biology over the years, I am going to assume that you at least know something about human chromosomes and perhaps other mammals. If you don’t, you may need to do a quick google review before you go any further. If you find this intimidating or overwhelming, I will give you the “skinny” on it. You can always review it later.
Here is how a typical textbook depicts a mammalian chromosome:
Looks like four weiners stuck together. If you check more closely, the textbook will indicate that each “weiner” is composed of a single, tightly wound, continuous thread of DNA covered with other materials like proteins. Such a complex is called chromatin. This thread travels from one short weiner to the adjacent long weiner. The weiners on the bottom are considered as “copies” of the ones on top as long as sex isn’t involved. The two weiners (long and short), together with their copies are connected together in the middle by a structure called a centromere.
There is a particular kind of chromosome called a lampbrush chromosome discovered back in the 1880’s. You can get a good idea of what they look like using the Wiki link above. These chromosomes have loops of DNA sticking out from them, making them look like old-fashioned lampbrushes. It is generally assumed that these loops are merely part of a continuous structure of DNA making up the entire chromosome. However, they are not randomly produced and have defined fixed locations within the chromosome. Studies in the late 1970’s show such loops appear to be universal among all kinds of chromosomes. A good source for understanding the relation of DNA loops to global chromosome structure can be found in the following 2002 NIH reference link. These loops behave like individual DNA domains, as if they were separate chromosomes themselves. Such an arrangement indicates complex temporal coordination of gene expression. In other words, these DNA domains cooperate with one another for the greater good of the cell. How these structures may have come to be part of our chromosomes is the main subject of this entire blog: smaller chromosomes building bigger chromosomes.
Bacterial chromosomes are generally circular in shape and have a wide range of sizes. A circle with a circumference of 340 microns (millionth of a meter) would be in the lower size range. Breaking this circle and laying it out end to end gives you a linear length of 340 microns which is 34 times the diameter of an average human nucleus. There is more than a million microns of DNA within this nucleus. To give you some idea of this magnitude, check out the graphic below:
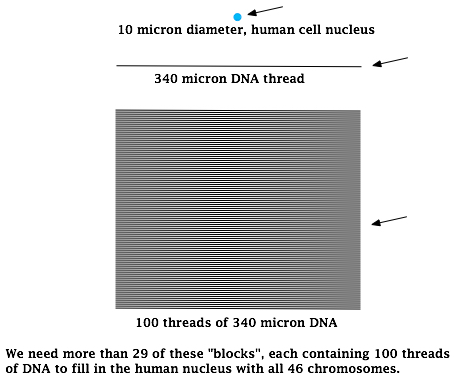
We need more than 29 of these “blocks”, each containing 100 threads of DNA to fill in the human nucleus with all 46 chromosomes.
That’s a lot of DNA to fit inside that tiny little nucleus, don’t you think? However, according to textbooks, all of that DNA is fused together into just 46 pieces of “rope”called chromosomes. Now imagine a piece of DNA like the one shown above breaking off or replicating from the main strand of a relatively small human chromosome, say one that is only 1700 times the length of the diameter of the nucleus. Now imagine this piece of DNA somehow coming back on itself and ligating the ends to form a 340 micron circle. Not much wiggle room is there? Even if there was, the chances of this meandering 340 micron thread of DNA spontaneously doing this is pretty remote at best. Yet, somehow it happens. Textbooks indicate that pieces of DNA the size of bacterial chromosomes can form into small circles within the human nucleus. You can learn more about this under general postings.
Wouldn’t it make more sense if that piece of DNA was circular to begin with? Imagine again, circles of DNA attached to other circles like grapes on a vine, easily plucked off from the main body. The photomicrograph shown above shows exactly this, only the grapes are beaded circles. Could these circles be a plucked version of lamp brush loops?
You would think scientists would be jumping all over this, wouldn’t you? You would be wrong. Indeed, the greatest mystery of all may be why they are NOT jumping all over this. After all, it has been 28 years since they were first discovered, and still nothing crops up in the literature. Now why is that?
You will find more about this under Blog #2. Suffice it to say, when a well-respected scientist tells you privately that “they will kill you for this!”, something is definitely rotten in Denmark, don’t you think?
Well, the stakes are high here. Many people are completely invested in a linear DNA human chromosome model, even to the point of sweeping aside discoveries such as these. Just check the literature and you’ll see what I mean. So in the great scheme of things, should it really matter to you how a human chromosome is put together? Well, only if you want to expand our knowledge of genetic diseases, aging, human development, and cancer, among other things.
You realize this doesn’t have to be the end of this. I know there are some very wealthy philanthropists out there looking for a worthy cause. All they have to do is contact me to learn more and how to move it forward. If you know anyone like that, please ask them to contact me.
Questions and comments are welcome.
Best regards,
Dr. Frank Abernathy
————————————
If you are a DNA biologist, please continue reading…
Let me boil things down about this blog as best as I can in as little space as possible. I believe the following things about chromosomes:
They are comprised of interconnected circles of chromatin joined together at their origins of replication. These circles are capable of fusing together during differentiation or completely separating, resulting in the loss of DNA circles.
Paired origins of replication from adjacent replicon circles of DNA give rise to promoters of transcription by fusing together and losing DNA vital to replication.
Paired origins of replication give rise to enhancers of transcription and/or replication when they separate from one another and one of them is discarded.
Paired origins of replication from adjacent replicon circles of DNA give rise to splice sites by fusing together and losing DNA vital to both replication and transcription.
These processes occur through phylogeny and ontogeny. In other words, these events occurred eons ago (ancestral) or they occur only when the embryo begins to differentiate into tissues (species specific).
You will understand what I am saying more clearly if you click on this video link. It is a roughly 40 minute presentation.
Here is some preliminary evidence to support my hypotheses. There are many additional references within this blog, my website, dissertation, and manuscripts. I would appreciate any additional information you can provide to help me fill out the table below:

1a) http://www.cell.com/ajhg/abstract/S0002-9297(07)62467-7 (2001)AT rich palindromes are associated with translocations
1b) http://hmg.oxfordjournals.org/content/10/23/2605.full (2001)Long AT-rich palindromes and the constitutional t(11;22) breakpoint
2) http://www.pnas.org/content/79/2/381.full.pdf (1982)SV 40 origins of replication missing AT rich regions within the palindrome could not replicate
3) http://genesdev.cshlp.org/content/2/9/1115 (1988)Enhancers required for replication once an embryonic nucleus is formed in mice.
4) http://nar.oxfordjournals.org/content/16/23/11207 (1988)Do transcriptional enhancers also augment DNA replication?
5) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC400889/ (1989)Replication origins can act as enhancers for amplification of other origins in Drosophila.
6) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4403523/ (2015)multiple origins of replication in bacteria. Double origins of replication.
7) http://www.ncbi.nlm.nih.gov/pubmed/9453148 (1997)TATA box in origin of papillomavirus 18 and requires and enhancer of replication
8) http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1000454 (2009)CpG Islands: Starting Blocks for Replication and Transcription
9) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC401248/ (1989) An enhancer located in a CpG-island 3′ to the TCR/CD3-epsilon gene confers T lymphocyte-specificity to its promoter.
10)http://www.sciencedirect.com/science/article/pii/S0014579303010925 (2003) Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity
11) http://www.ncbi.nlm.nih.gov/pubmed/12704365 (2003)Characterization of a palindromic enhancer element in the promoters of IL4, IL5, and IL13 cytokine genes.
12) http://www.genomebiology.com/2004/5/12/251 (2004) The origin of recent introns: transposons?
14) Origin pairing (‘handcuffing’) as a mode of negative control of P1 plasmid copy number. (2001)

You made some nice points there. I did a search on the topic and found most guys will approve with your site.
Awesome stuff you guys got here.I really like the theme of the website and how well you organized the content. It’s a marvelous job I will come back and check you out sometime,come on over to my site and i will show you how to make a full time living with your blog without selling anything using the internet’s most powerful marketing scraping and posting software “scrapebox”
Throughout the grand pattern of things you get an A+ with regard to hard work. Where exactly you actually lost everybody was first in your facts. You know, it is said, details make or break the argument.. And that couldn’t be more true right here. Having said that, allow me say to you what did give good results. Your authoring is actually pretty convincing and this is most likely the reason why I am making the effort to comment. I do not make it a regular habit of doing that. 2nd, despite the fact that I can easily notice a leaps in reasoning you make, I am not convinced of just how you seem to connect your points which inturn help to make the actual conclusion. For the moment I shall yield to your issue however wish in the future you actually connect the facts much better.
I believe you have mentioned some very interesting details , thanks for the post.
I really appreciate this post. I have been looking everywhere for this! Thank goodness I found it on Bing. You have made my day! Thx again!
Unquestionably believe that which you stated.
Your favorite justification appeared to be on the internet the easiest thing to be aware of.
I say to you, I certainly get annoyed while people think about worries
that they just don’t know about. You managed to
hit the nail upon the top as well as defined out the whole thing without having side-effects , people could take a signal.
Will probably be back to get more. Thanks
So sorry for such a late response to your comment. I can see why you find some of my conclusions as giant leaps of faith. Unfortunately, there are no shortcuts here. To gain any depth in how I come up with my hypotheses requires perusing not only my blog but also many of the papers which are cited here. This is why I started up a series of Bitchute videos: https://www.bitchute.com/channel/1nKdCzP3wq0M/. I think your best bet would be to look at my most recent Bitchute video which provides a visual interpretation of my “ARR” manuscripts. These manuscripts can be found right here on my blog and they have been downloaded numerous times. They are quite technical in nature, hence the use of a video to boil it all down for the casual visitor. Of course, you can always ask me any questions you may have about any of the content in this blog, videos, or literature via e mail: fabernathy@sbcglobal.net. In fact, I have posted a transcript of a “DNA interview” I had with a “reporter” right here on this blog. You can locate it on the main menu bar page tabs above this blog.
I’ve been browsing on-line greater than three hours these days, yet I by no means found any attention-grabbing article like yours. It is pretty price enough for me. Personally, if all website owners and bloggers made good content material as you did, the net will likely be much more useful than ever before.
Hi there! Someone in my Myspace group shared this website with us so I came to give it a look. I’m definitely loving the information. I’m bookmarking and will be tweeting this to my followers! Excellent blog and wonderful style and design.
I am impressed with this web site , really I am a big fan .
Hello! I simply would like to offer you a huge thumbs up for your great info
you’ve got here on this post. I’ll be coming back to your website for more soon.
Thanks for the blog.Thanks Again. Really Cool.
I think this is a real great blog.Thanks Again. Really Great.
Very neat post.Thanks Again. Much obliged.
Wow, great blog.Much thanks again. Really Cool.
I really like and appreciate your article. Fantastic.
Really appreciate you sharing this blog post.Really thank you! Will read on…
Thanks-a-mundo for the article.Thanks Again. Awesome.
Really enjoyed this post.Thanks Again. Will read on…
I am so grateful for your article. Want more.
Great, thanks for sharing this blog post.Much thanks again. Cool.
I value the blog article. Cool.
Major thanks for the blog.Really looking forward to read more. Keep writing.
Im obliged for the post.Really looking forward to read more. Fantastic.
I really like and appreciate your blog article.Really thank you! Great.
I truly appreciate this blog post.Really thank you!
wow, awesome article.Really thank you! Much obliged.
Im grateful for the article.Much thanks again.
Very informative post.Really thank you! Really Great.
Thanks a lot for the article post.Much thanks again. Awesome.
Major thankies for the blog.Really looking forward to read more. Keep writing.
Great article.Much thanks again. Fantastic.
Thanks a lot for the blog article.Really looking forward to read more. Really Great.
A round of applause for your blog post.Really thank you! Will read on…
I loved your blog post.Really looking forward to read more. Cool.
What’s Taking place i’m new to this, I stumbled upon this I have discovered It positively helpful and it has helped me out loads. I hope to contribute & aid other users like its aided me. Great job.
Great article.Really thank you! Want more.
I really liked your blog. Really Great.
Great blog article.Much thanks again. Really Cool.
Really enjoyed this article post.Really looking forward to read more.
Great, thanks for sharing this blog article.
A round of applause for your article post.Really thank you! Awesome.
I am so grateful for your blog post. Fantastic.
Major thankies for the blog article. Really Cool.
Hmm it seems like your blog ate my first comment (it was extremely long) so I guess I’ll just sum it up what I submitted and say, I’m thoroughly enjoying your blog. I as well am an aspiring blog writer but I’m still new to everything. Do you have any recommendations for beginner blog writers? I’d definitely appreciate it.
I appreciate you sharing this post.Much thanks again. Really Great.
Great post, you have pointed out some fantastic details , I besides believe this s a very great website.
I do accept as true with all of the ideas you have presented for your post. They are really convincing and will certainly work. Nonetheless, the posts are very quick for newbies. May you please extend them a bit from subsequent time? Thank you for the post.
Really informative blog article. Fantastic.
Appreciate you sharing, great post.Really thank you! Fantastic.
I cannot thank you enough for the article.Much thanks again. Awesome.
Thanks-a-mundo for the blog post. Much obliged.
Enjoyed every bit of your post. Keep writing.
Really enjoyed this blog. Will read on…
Keep functioning ,remarkable job!
Super-Duper site! I am loving it!! Will be back later to read some more. I am taking your feeds also.
I’m still learning from you, as I’m trying to achieve my goals. I absolutely enjoy reading all that is written on your site.Keep the information coming. I liked it!
Good day! This is my first comment here so I just wanted to give a quick shout out and say I genuinely enjoy reading through your articles. Can you suggest any other blogs/websites/forums that go over the same subjects? Many thanks!
I do not even know how I ended up here, but I thought this post was good. I don’t know who you are but certainly you are going to a famous blogger if you are not already 😉 Cheers!
very nice put up, i actually love this web site, keep on it
I’m no longer certain where you are getting your information, however good topic. I must spend some time learning more or working out more. Thanks for wonderful info I used to be searching for this information for my mission.
Hello there, I discovered your blog by way of Google at the same time as searching for a related topic, your website came up, it looks great. I’ve bookmarked it in my google bookmarks.
Very neat article post.Thanks Again. Fantastic.
Nice post. I learn something more challenging on totally different blogs everyday. It’ll always be stimulating to learn content material from different writers and follow a little bit one thing from their store. I’d choose to make use of some with the content on my blog whether or not you don’t mind. Natually I’ll give you a hyperlink on your internet blog. Thanks for sharing.
Thanks a lot for the blog. Want more.
I really like what you guys tend to be up too. This sort of clever work and reporting! Keep up the awesome works guys I’ve incorporated you guys to blogroll.
You made some decent points there. I looked on the internet for the problem and found most people will go together with with your website.
A round of applause for your post.Much thanks again. Awesome.
Hello there, You have done a great job. I’ll certainly digg it and personally suggest to my friends. I’m confident they will be benefited from this website.
Woah! I’m really digging the template/theme of this site. It’s simple, yet effective. A lot of times it’s very difficult to get that “perfect balance” between superb usability and visual appeal. I must say that you’ve done a superb job with this. Additionally, the blog loads super quick for me on Chrome. Superb Blog!
Major thankies for the article.Really thank you! Really Great.
I do consider all the concepts you have offered to your post. They are very convincing and can certainly work. Still, the posts are very quick for novices. May you please lengthen them a little from next time? Thanks for the post.
I think this is a real great post.Thanks Again. Cool.
I truly appreciate this blog article.Much thanks again. Great.
Awesome blog article.Much thanks again. Really Cool.
I appreciate you sharing this post. Keep writing.
Thanks for the sensible critique. Me and my neighbor were just preparing to do some research on this. We got a grab a book from our area library but I think I learned more clear from this post. I’m very glad to see such great information being shared freely out there.
Bloghopping is really my forte and i like to visit blogs,
Enjoyed every bit of your blog article.Much thanks again. Cool.
Usually I don’t read article on blogs, but I would like to say that this write-up very forced me to try and do it! Your writing style has been amazed me. Thanks, quite nice post.
Great news once again!
Im obliged for the article post.Thanks Again. Fantastic.
Amazin!
Wow! Dies könnte einer der nützlichsten Blogs sein, die wir je zu diesem Thema gefunden haben. Grundlegend Grundsätzlich Großartig Wundervoll Fantastisch Hervorragend. Ich bin auch ein Experte auf diesem Gebiet, deshalb kann ich Ihre harte Arbeit verstehen.
Translation: Wow! This could be one of the most useful blogs we have ever found on the subject. Basic Basic Great Wonderful Fantastic Outstanding. I am also an expert in this field so I can understand your hard work.
Major thankies for the article post. Really Cool.
I am so grateful for your article. Fantastic.
I’d need to test with you here. Which isn’t something I often do! I take pleasure in reading a post that may make folks think. Additionally, thanks for allowing me to comment!
I loved your blog.Thanks Again. Want more.
Heya i’m for the first time here. I found this board and I find It truly useful & it helped me out a lot. I hope to give something back and help others like you aided me.
Thank you for your blog.Really looking forward to read more. Cool.
Thanks a lot for the blog.Really looking forward to read more. Really Great.
Im grateful for the post.Much thanks again. Cool.
Thank you for another fantastic article. Where else may just anyone get that kind of information in such an ideal way of writing? I have a presentation next week, and I am on the look for such info.
I really liked your blog article.Much thanks again. Awesome.
This is one awesome blog post. Keep writing.
Appreciate you sharing, great article post.Really looking forward to read more. Great.
What’s Happening i am new to this, I stumbled upon this I’ve found It positively helpful and it has helped me out loads. I hope to contribute & help other users like its helped me. Great job.
This is one awesome blog.Really thank you! Awesome.
Im obliged for the article.Much thanks again. Really Great.
Wow! Thank you! I continually needed to write on my blog something like that. Can I implement a part of your post to my site?
I truly appreciate this post.Thanks Again. Will read on…
Really enjoyed this blog post.Really thank you! Want more.
It’s difficult to find knowledgeable people on this topic, but you seem like you know what you’re talking about! Thanks
Hi there! Do you use Twitter? I’d like to follow you if that would be ok. I’m undoubtedly enjoying your blog and look forward to new updates.
I love reading your website.
Wonderful article! We are linking to this particularly great content on our site. Keep up the great writing.
I truly appreciate this post. I have been looking everywhere for this! Thank goodness I found it on Bing. You’ve made my day! Thanks again
You made some good points there. I checked on the net to find out more about the issue and found most people will go along with your views on this site.
Everyone loves it when individuals get together and share thoughts. Great website, stick with it!
Very informative post. Will read on…
Hello there, You’ve done a great job. I’ll definitely digg it and personally suggest to my friends. I’m confident they’ll be benefited from this web site.
Thanks so much for the article. Cool.
Looking forward to reading more. Great blog article. Cool.
naturally like your website but you have to check the spelling on several of your posts. A number of them are rife with spelling issues and I find it very bothersome to tell the truth nevertheless I’ll certainly come back again.
Im obliged for the blog article.Much thanks again. Fantastic.
Rife with spelling issues? Are you certain you have the right blog? If you don’t mind, please point out some examples for me to correct. Which page, which post, which paragraph? Thanks again.
Oh my goodness! Impressive article dude! Thank you so much, However I am encountering troubles with your RSS. I don’t know why I can’t subscribe to it. Is there anyone else getting similar RSS issues? Anyone that knows the answer will you kindly respond? Thanx!!
Amazing content here.
Hi, I do believe this is a great site. I stumbledupon it 😉 I’m going to revisit yet again since i have book-marked it. Money and freedom is the best way to change, may you be rich and continue to help other people.
Your style is unique compared to other people I’ve read stuff from. Thanks for posting when you’ve got the opportunity, Guess I will just bookmark this blog.
Hi there! I simply want to offer you a huge thumbs up for the great information you’ve got right here on this post. I am returning to your website for more soon.
You’re so cool! I do not suppose I’ve read anything like this before. So great to find another person with a few genuine thoughts on this subject. Seriously.. many thanks for starting this up. This website is something that is needed on the internet, someone with some originality!
Hi, I do believe this is an excellent web site. I stumbledupon it 😉 I will revisit once again since I book-marked it. Money and freedom is the greatest way to change, may you be rich and continue to help others.
There’s certainly a great deal to know about this topic. I really like all of the points you’ve made.
Thanks again for the blog post.Much thanks again. Great.
Very neat blog article.Much thanks again. Awesome.
This site certainly has all of the info I wanted concerning this subject and didn’t know who to ask.
I must thank you for the efforts you’ve put in penning this site. I’m hoping to check out the same high-grade blog posts from you in the future as well. In truth, your creative writing abilities has encouraged me to get my own blog now 😉
I seriously love your website.. Excellent colors & theme. Did you make this amazing site yourself? Please reply back as I’m wanting to create my own personal site and would like to learn where you got this from or what the theme is named. Appreciate it!
Good post. I learn something totally new and challenging on blogs I stumbleupon on a daily basis. It will always be exciting to read content from other authors and practice something from other sites.
I blog frequently and I truly appreciate your content. Your article has truly peaked my interest. I will take a note of your site and keep checking for new information about once a week. I opted in for your Feed as well.
Dear Fellow Friend, Liking the persistence you put into your blog and extensive information you provide.
I blog often and I genuinely thank you for your information. This article has truly peaked my interest. I will take a note of your site and keep checking for new information about once a week. I opted in for your Feed as well.
This is a topic which is near to my heart… Cheers! Exactly where are your contact details though?
Great information. Lucky me I recently found your blog by accident (stumbleupon). I have bookmarked it for later!
Very neat post.Really looking forward to read more. Will read on…
Check the page tab called “contact Form”.
Your style is unique in comparison to other folks I’ve read stuff from. Thank you for posting when you’ve got the opportunity, Guess I will just book mark this blog.
Thank you ever so for you article.Thanks Again.
This is one awesome blog article.Thanks Again. Fantastic.
I blog often and I seriously thank you for your content. This great article has truly peaked my interest. I am going to book mark your site and keep checking for new information about once a week. I opted in for your RSS feed too.
Good information. Lucky me I ran across your website by chance (stumbleupon). I have saved as a favorite for later!
You are a very smart person!
I really like it when folks get together and share views. Great website, continue the good work!
I cannot thank you enough for the post.Really looking forward to read more. Much obliged.
Your style is really unique compared to other folks I have read stuff from. I appreciate you for posting when you have the opportunity, Guess I will just bookmark this site.
Wow, great blog article.Much thanks again. Great.
I just want to tell you that I’m all new to blogs and definitely liked you’re web page. Almost certainly I’m want to bookmark your website . You definitely come with perfect articles and reviews. Many thanks for sharing with us your web-site.
I need to to thank you for this fantastic read!! I certainly loved every little bit of it. I have you book marked to check out new stuff you post…
I’m extremely inspired along with your writing talents as smartly as with the structure for your weblog. Is that this a paid subject matter or did you modify it yourself? Anyway stay up the nice high quality writing, it is uncommon to peer a nice weblog like this one today..
I think other web-site proprietors should take this web site as an model, very clean and magnificent user genial style and design, let alone the content. You are an expert in this topic!
I truly appreciate this article post.Really thank you! Fantastic.
Thanks-a-mundo for the blog article. Awesome.
Hello! I simply would like to offer you a big thumbs up for your great information you’ve got here on this post. I will be coming back to your site for more soon.
Hello. magnificent job. I did not imagine this. This is a remarkable story. Thanks!
Pretty! This was a really wonderful article. Many thanks for supplying this information.
Excellent article! We will be linking to this particularly great article on our site. Keep up the good writing.
I’m extremely pleased to discover this site. I want to to thank you for your time due to this wonderful read!! I definitely appreciated every little bit of it and i also have you book-marked to check out new stuff on your website.
Good day! I just want to offer you a huge thumbs up for your great info you’ve got here on this post. I am coming back to your blog for more soon.
Saved as a favorite, I really like your site!
There is definately a great deal to know about this subject. I love all the points you’ve made.
You’re so interesting! I do not believe I’ve truly read through anything like this before. So nice to find someone with some unique thoughts on this topic. Really.. thank you for starting this up. This website is one thing that is required on the web, someone with a bit of originality!
Thank you.
I could not refrain from commenting. Well written!
Good day! I could have sworn I’ve been to this blog before but after looking at a few of the posts I realized it’s new to me. Anyways, I’m certainly happy I found it and I’ll be book-marking it and checking back often!
Spot on with this write-up, I seriously feel this website needs much more attention. I’ll probably be back again to see more, thanks for the information!
It’s exhausting to seek out knowledgeable folks on this subject, but you sound like you know what you’re speaking about! Thanks
I really liked your blog article.
Spot on with this write-up, I actually think this site needs far more attention. I’ll probably be back again to see more, thanks for the info!
This site was… how do you say it? Relevant!! Finally I’ve found something which helped me. Appreciate it!
Hello there! This post could not be written much better! Looking through this article reminds me of my previous roommate! He continually kept talking about this. I’ll send this article to him. Fairly certain he will have a great read. Thank you for sharing!
Aw, this was a really nice post. Spending some time and actual effort to make a superb article… but what can I say… I put things off a whole lot and never manage to get nearly anything done.
After looking over a handful of the blog posts on your web page, I truly like your technique of writing a blog. I saved it to my bookmark website list and will be checking back in the near future. Take a look at my website too and let me know your opinion.
Hey there! I just would like to offer you a huge thumbs up for your excellent info you’ve got right here on this post. I am coming back to your blog for more soon.
I’m very pleased to discover this page. I need to to thank you for your time for this fantastic read!! I definitely liked every part of it and I have you saved as a favorite to check out new information on your website.
There is certainly a great deal to find out about this topic. I really like all of the points you have made.
Appreciate you sharing, great blog.Thanks Again. Keep writing.
I needed to thank you for this great read!! I certainly enjoyed every bit of it. I’ve got you bookmarked to check out new stuff you post…
I was able to find good advice from your content.
Good post! We are linking to this great article on our website. Keep up the great writing.
Having read this I believed it was rather informative. I appreciate you taking the time and energy to put this article together. I once again find myself personally spending way too much time both reading and posting comments. But so what, it was still worth it!
Aw, this was a really nice post. Taking a few minutes and actual effort to make a great article… but what can I say… I procrastinate a whole lot and don’t seem to get nearly anything done.
bookmarked!!, I love your web site!
Enjoyed every bit of your blog.Really looking forward to read more. Fantastic.
I blog frequently and I truly appreciate your information. Your article has truly peaked my interest. I am going to bookmark your site and keep checking for new details about once per week. I subscribed to your Feed as well.
Excellent web site you have got here.. It’s hard to find high quality writing like yours nowadays. I really appreciate individuals like you! Take care!!
This website definitely has all of the info I needed concerning this subject and didn’t know who to ask.
Good site you have got here.. It’s hard to find excellent writing like yours nowadays. I really appreciate people like you! Take care!!
Fantastic post.Really thank you! Want more.
Awesome article.Much thanks again. Fantastic.
Aw, this was a really nice post. In concept I wish to put in writing like this moreover – taking time and precise effort to make an excellent article… however what can I say… I procrastinate alot and under no circumstances seem to get one thing done.
This website was… how do I say it? Relevant!! Finally I have found something that helped me. Kudos!
A big thank you for your blog post.Really looking forward to read more. Will read on…
Thanks for sharing, this is a fantastic blog.Really looking forward to read more. Awesome.
Thanks for the marvelous posting! I quite enjoyed reading it, you could be a great author.I will be sure to bookmark your blog and definitely will come back very soon. I want to encourage you to continue your great writing, have a nice weekend!
Great information. Lucky me I came across your site by accident (stumbleupon). I’ve bookmarked it for later!
I am really inspired with your writing talent well with the layout to your weblog. Is this a paid topic or did you modify it your self? Anyway stay up the excellent high quality writing, it’s rare to peer a great blog like this one nowadays.
bookmarked!!, I like your website!
Saved as a favorite, I like your blog!
This is a free blog about the research I did at Ohio State University. You might like my Bitchute video channel too: When it Chromosomes,You Have Been Misled: http://www.bitchute.com/channel/1nKdCzP3wq0M/
I’m typically to blogging and i actually respect your content. The article has really peaks my interest. I am going to bookmark your site and maintain checking for brand spanking new information.
Pretty great post. I just stumbled upon your blog and wished to say that I’ve really loved surfing around your blog posts. In any case I’ll be subscribing for your rss feed and I’m hoping you write once more very soon!
Hi! I could have sworn I’ve visited this site before but after browsing through some of the posts I realized it’s new to me. Regardless, I’m definitely happy I found it and I’ll be bookmarking it and checking back often!
Hello! I just would like to give a huge thumbs up for the great info you have here on this post. I will be coming back to your blog for more soon.
I am so grateful for your blog article.Really thank you! Awesome.
Spot on with this write-up, I actually believe that this website needs far more attention. I’ll probably be returning to see more, thanks for the advice!
An interesting discussion is definitely worth comment. I do believe that you ought to publish more on this topic, it may not be a taboo matter but typically people don’t discuss these subjects. To the next! Best wishes!!
Hi there! This blog post could not be written much better! Looking at this post reminds me of my previous roommate! He continually kept preaching about this. I most certainly will send this article to him. Pretty sure he’ll have a very good read. Thanks for sharing!
Wow, great article post. Cool.
Saved as a favorite, I like your website!
You need to take part in a contest for one of the most useful sites online. I’m going to recommend this blog!
Pretty! This was an extremely wonderful post. Thanks for providing these details.
Thanks for sharing superb informations. Your website is so cool. I’m impressed by the details that youÃve on this website. It reveals how nicely you perceive this subject. Bookmarked this website page, will come back for extra articles. You, my pal, ROCK! I found simply the information I already searched all over the place and just couldn’t come across. What a great site.
Aw, this was an exceptionally nice post. Spending some time and actual effort to generate a good article… but what can I say… I hesitate a lot and never seem to get anything done.
I’d like to thank you for the efforts you’ve put in writing this blog. I am hoping to check out the same high-grade blog posts by you in the future as well. In fact, your creative writing abilities has motivated me to get my own blog now 😉
Good blog you have got here.. It’s hard to find excellent writing like yours nowadays. I honestly appreciate individuals like you! Take care!!
Very good post. I absolutely appreciate this site. Thanks!
Very good blog post.Thanks Again. Really Great.
This is one awesome article. Keep writing.
This site was… how do I say it? Relevant!! Finally I have found something which helped me. Thanks a lot!
Major thankies for the blog.Thanks Again. Fantastic.
Very good info. Lucky me I recently found your website by chance (stumbleupon). I’ve book-marked it for later!
Hi there! This blog post could not be written any better! Looking through this post reminds me of my previous roommate! He constantly kept talking about this. I’ll forward this information to him. Fairly certain he’ll have a great read. I appreciate you for sharing!
Major thanks for the post.Thanks Again.
Hi, I think your blog might be having browser compatibility issues. When I look at your blog site in Ie, it looks fine but when opening in Internet Explorer, it has some overlapping. I just wanted to give you a quick heads up! Other then that, very good blog!
Thanks on your marvelous posting! I actually enjoyed reading it, you can be a great author. I will always bookmark your blog and will often come back from now on. I want to encourage yourself to continue your great job, have a nice evening!
You need to take part in a contest for one of the best blogs on the web. I’m going to highly recommend this web site!
I needed to thank you for this great read!! I definitely loved every little bit of it. I’ve got you book-marked to check out new stuff you post…
Pretty! This was an incredibly wonderful article. Thank you for providing these details.
Hello! I simply want to offer you a huge thumbs up for your excellent information you’ve got right here on this post. I will be returning to your web site for more soon.
This is a topic that is close to my heart… Thank you! Where are your contact details though?
Try the contact page on the menu bar.
Hello There. I found your blog using msn. This is an extremely well written article. I will be sure to bookmark it and return to read more of your useful information. Thanks for the post. I’ll certainly comeback.
This is a fantastic web page, could you be interested in doing an interview about just how you designed it? If so e-mail me!
Really informative blog article.Much thanks again. Keep writing.
Necessary to send you a little note to help Thanks much again on the magnificent views that you have discussed at this time. It really is incredibly generous easily give you what exactly most people would have made as an e-book to get some bucks for themselves, especially considering that you could possibly have attempted in the event where you want. Similarly, the guidelines served to become a fantastic way to know that most people have identical to mine the same desire to learn much when considering this matter. I believe that thousands more fun times in the future for the people who look at your blog.
I responded to you via email.
Glad to be one of many visitors on this amazing web site : D.
Your style is so unique compared to other people I’ve read stuff from. Many thanks for posting when you’ve got the opportunity, Guess I will just bookmark this site.
when I imagine, it isn’t on daily basis that you’re ideal and it’s certainly not every single day that men and women notice ones exact model of the world… but this excellent posting was incredibly motivating… warm regards
I was more than happy to uncover this page. I want to to thank you for ones time for this wonderful read!! I definitely liked every bit of it and I have you saved as a favorite to check out new information in your web site.
Hi! I simply wish to give you a huge thumbs up for your excellent info you have here on this post. I will be coming back to your site for more soon.
Way cool! Some extremely valid points! I appreciate you penning this article and also the rest of the site is extremely good.
Absolutely outstanding information and very well written,thank you very much for this.
I appreciate you sharing this blog.Really looking forward to read more. Will read on…
I would really love to guest post on your blog.*~`~;
An interesting discussion is worth comment. There’s no doubt that that you ought to write more on this subject matter, it might not be a taboo matter but generally folks don’t talk about such issues. To the next! Many thanks!!
What would you like to post?
Great site you have here.. It’s hard to find high-quality writing like yours nowadays. I honestly appreciate individuals like you! Take care!!
Hi! I just would like to offer you a big thumbs up for the great info you’ve got here on this post. I’ll be returning to your website for more soon.
Loving the information on this internet site , you have done great job on the blog posts.
Normally I do not learn article on blogs, but I would like to say that this write-up very pressured me to try and do so! Your writing taste has been amazed me. Thank you, very great post.
Thanks for the marvelous posting! I certainly enjoyed reading it, you may be a great author. I will be sure to bookmark your blog and definitely will come back later in life. I want to encourage one to continue your great work, have a nice weekend!
You ought to take part in a contest for one of the finest sites on the internet. I am going to highly recommend this blog!
I blog quite often and I genuinely thank you for your information. This article has truly peaked my interest. I am going to take a note of your blog and keep checking for new information about once a week. I opted in for your RSS feed too.
I really like reading through a post that can make people think. Also, thank you for permitting me to comment!
It’s difficult to find experienced people on this subject, but you sound like you know what you’re talking about! Thanks
Howdy! I just wish to offer you a huge thumbs up for the great info you’ve got right here on this post. I will be returning to your website for more soon.
Saved as a favorite, I love your blog!
Thanks so much for the blog article.Much thanks again. Much obliged.
You’ve made some decent points there. I checked on the net for more information about the issue and found most individuals will go along with your views on this website.
Spot on with this write-up, I honestly think this site needs a great deal more attention. I’ll probably be returning to read through more, thanks for the info!
I am commenting to let you be aware of of the fabulous discovery my wife’s child gained going through the blog. She noticed a wide variety of issues, with the inclusion of what it is like to possess a great helping nature to get others very easily fully grasp a variety of extremely tough topics. You truly did more than readers’ expectations. I appreciate you for displaying such invaluable, trustworthy, educational as well as cool tips on this topic to Sandra.
I have to thank you for the efforts you have put in penning this site. I’m hoping to check out the same high-grade blog posts from you in the future as well. In fact, your creative writing abilities has motivated me to get my own blog now 😉
Hi! I just want to offer you a big thumbs up for your excellent info you’ve got right here on this post. I am coming back to your blog for more soon.
Did you have any fears or hesitations about meeting with a financial planner? My main hesitation was anxiety about seeing what our financial situation actually was, and also wondering if financial planning would be ‘worth it’ in terms of actionable advice.
Bertha Rand fought for decades with neighbours, police and the courts over keeping dozens of felines in her St. James home. Reporter Ben Waldman looks back at the life of Rand, who became an …
Sensational is your full time monitoring remedy that makes getting things done faster and also less complicated than ever before.
A big thank you for your article. Fantastic.